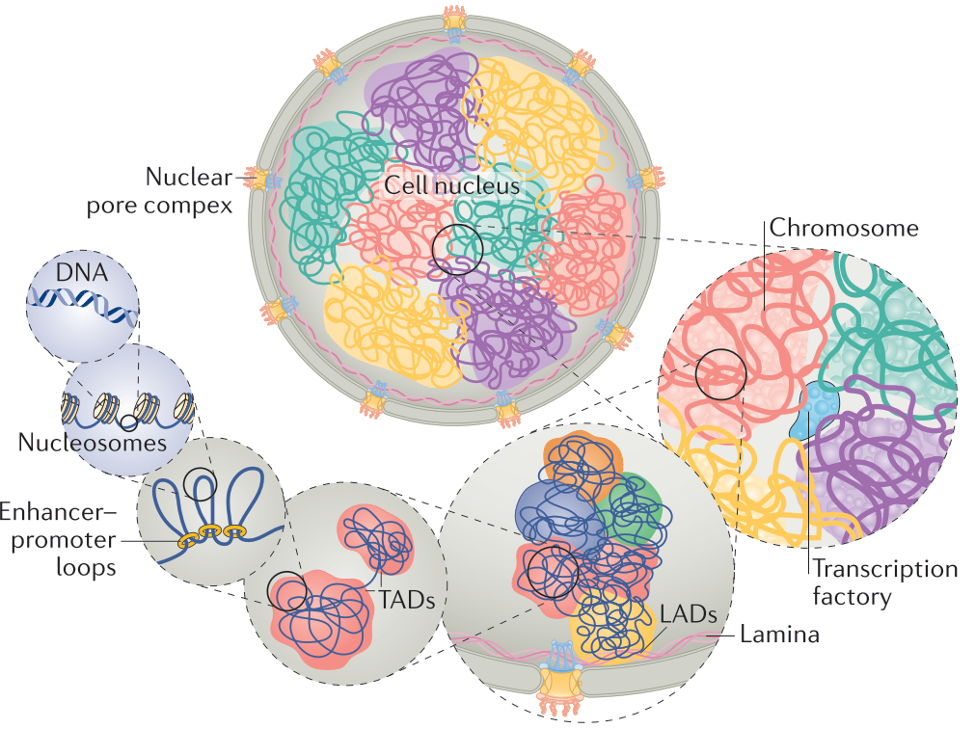
The invention relates to a sequencing method, able to detect chromatin changes, specifically heterocromatin. Alterations in heterochromatin are associated with developmental defects and cancer, while its proper conformation is a hallmark of healthy cells. As such, reliable methods to characterize heterochromatin and its alterations are essential for the biomedical scientific community.